Michael R. Garvin, Ph.D.
Founder & Chief Scientific Officer | Research Professor, University of New Mexico
Dr. Garvin is a molecular geneticist with over 20 years of experience in precision medicine and computational biology. He holds a Ph.D. from the University of Alaska Fairbanks and a Certificate in Personalized & Genomic Medicine from the University of Colorado Denver. His career spans Oak Ridge National Laboratory, Oregon State University, and biotechnology companies including Tularik (Amgen) and CV Therapeutics (Gilead).
His research focuses on identifying the genetic basis of complex diseases through novel computational approaches. With over 37 peer-reviewed publications in journals including Genome Biology, eLife, and PLoS Genetics, and over $5 million in NIH and DOE funding, Dr. Garvin has established himself as a leader in precision medicine research.
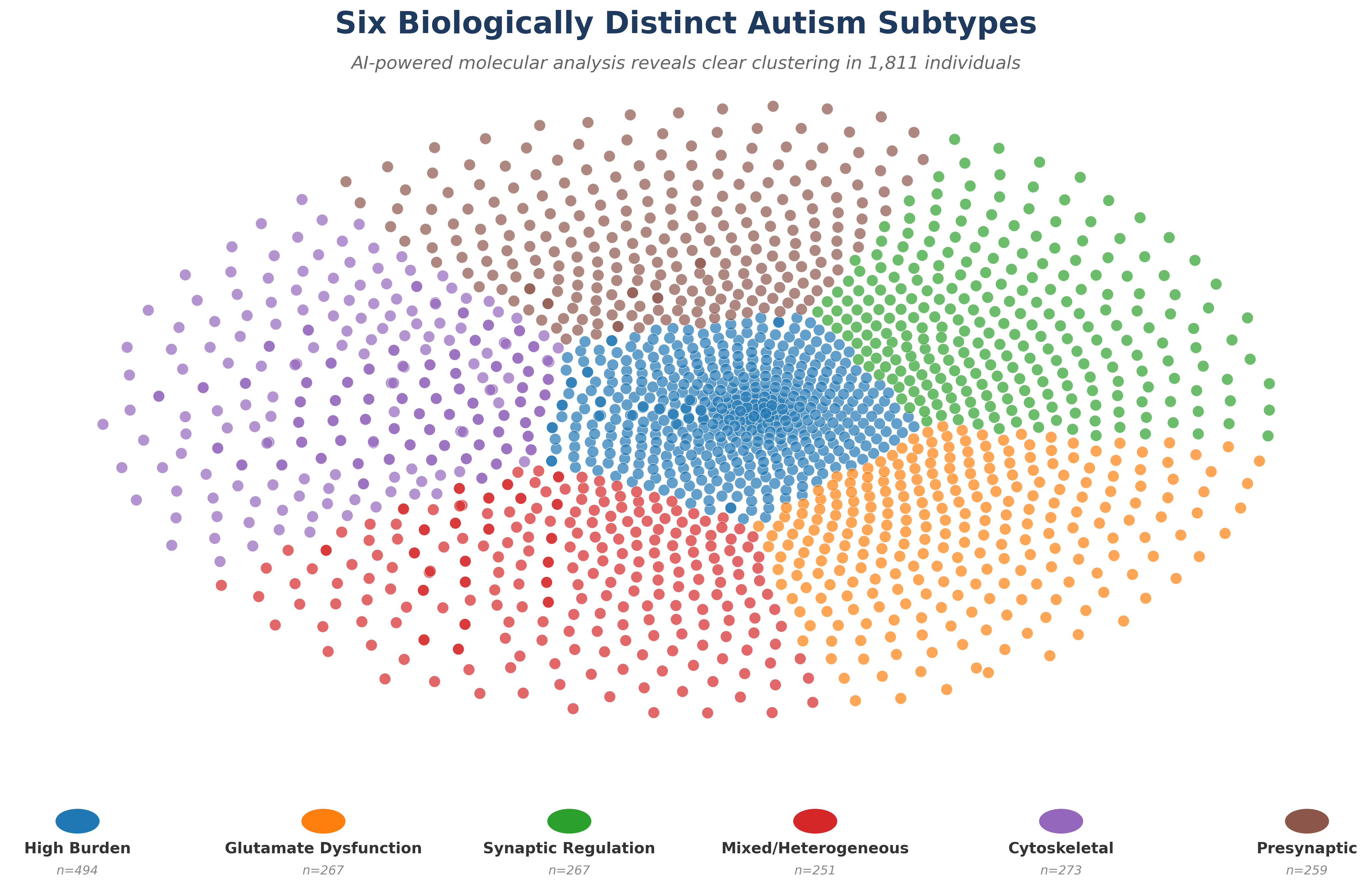
His COVID-19 research became the 2nd most viewed article in eLife history with over 154,000 views. Dr. Garvin founded Williwaw Biosciences after discovering that family-based genomic data contains rich subtyping information systematically discarded by conventional analysis pipelines—a breakthrough that enables precision medicine across any complex disease.